Identify the enriched biological themes
8. At the lower part of the list analysis page, you can find several enrichment widgets. Let’s start from the ‘Pathway Enrichment’ widget.
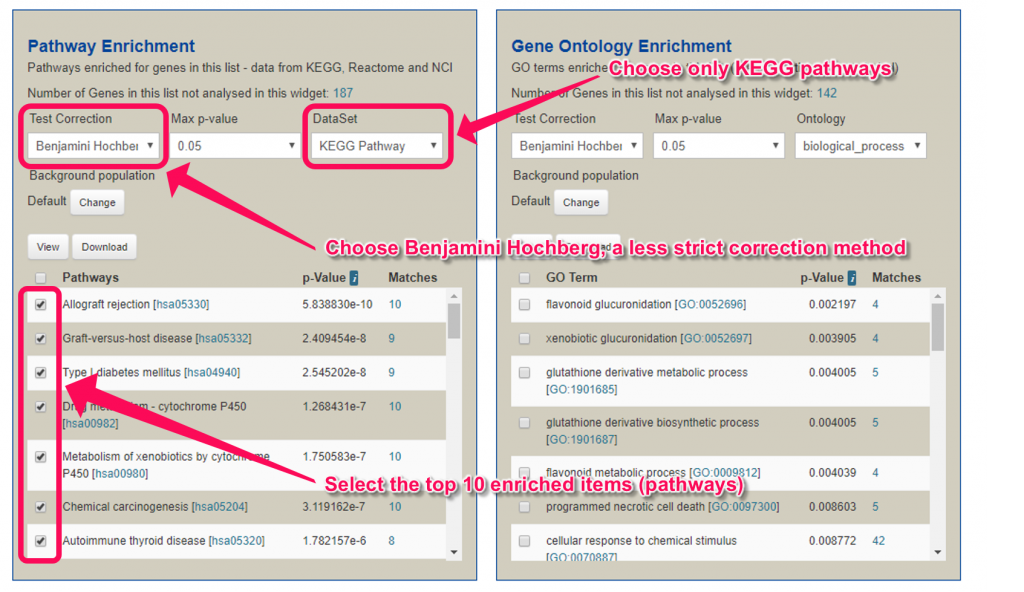
9. Change the ‘Test Correction’ to ‘Benjamini Hochberg’, which is a less strict multiple testing error correction method.
10. In this example, we only want to use KEGG pathway. Thus, change the ‘DataSet’ to ‘KEGG pathways data Set’.
11. Check the checkbox beside the top 10 enriched pathways (which has the least p-values).
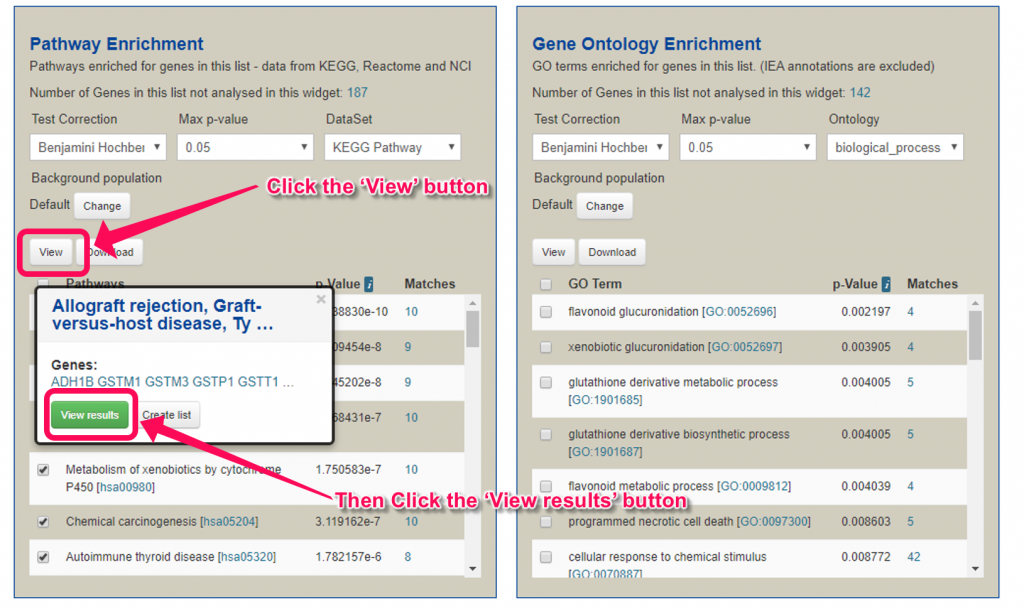
12. Click the ‘View’ button above the table, then click ‘View results’ button.
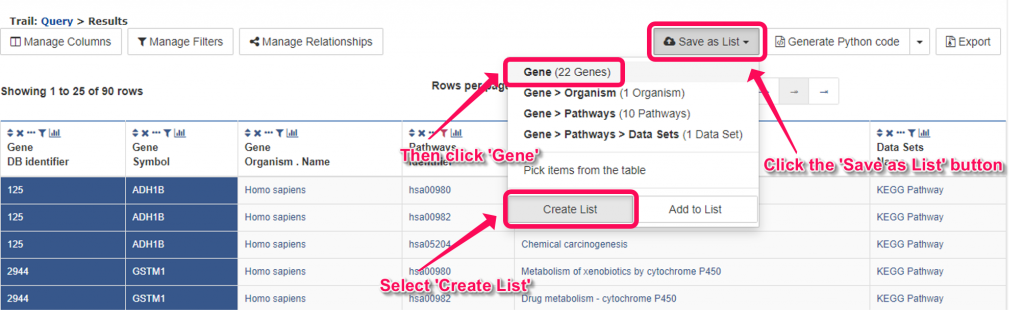
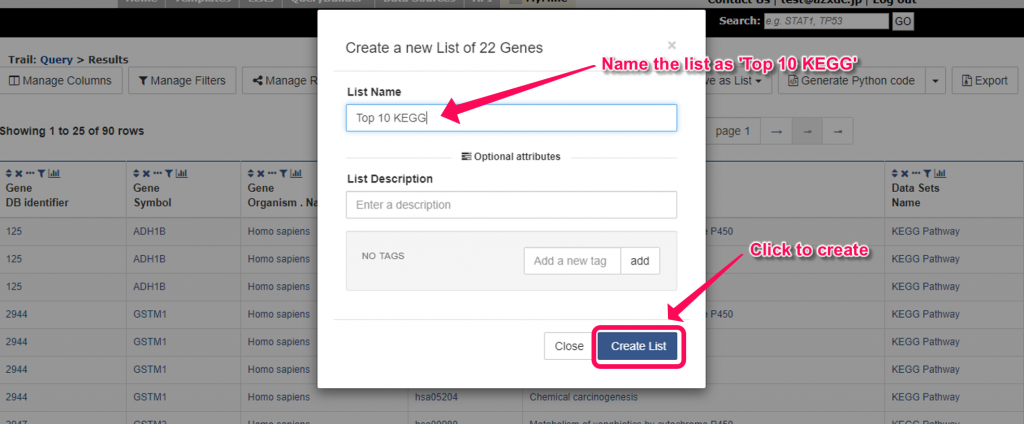
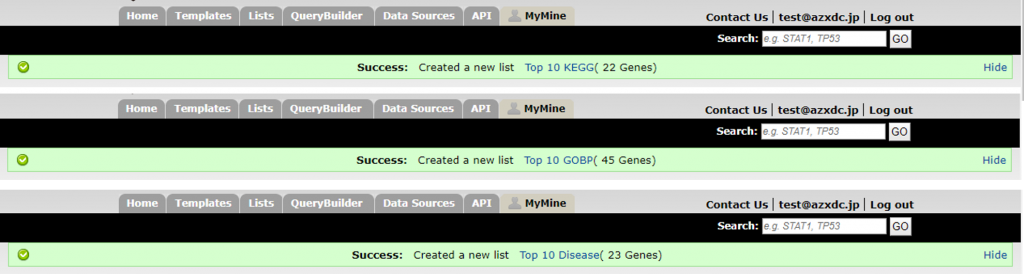
13. You will see the results in another tab. Click ‘Save as List’ button at the upper right of the table, then select ‘Create List’ and click ‘Gene’ on the menu. In the pop-up windows, enter ‘Top 10 KEGG’ as the name and click ‘Create List’ button. The genes in the list which associated with the top 10 enriched KEGG pathways will be saved as a new list named ‘Top 10 KEGG’.
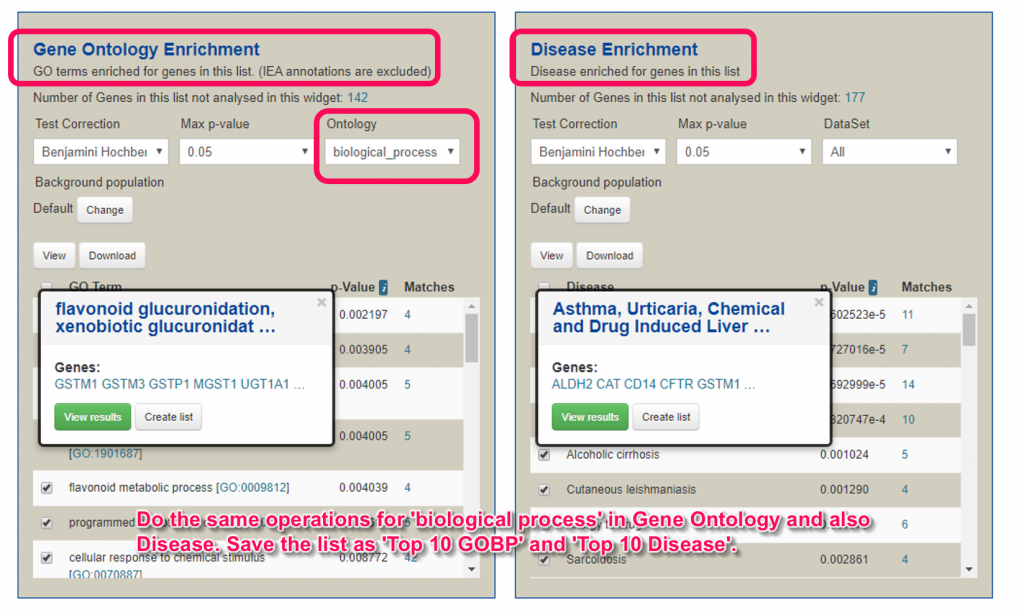
14. Do the same operations for GO and Disease to get 2 more gene lists. If you have no idea, here are descriptions for the operations.
- In the Gene Ontology Enrichment widget (if ‘Request Failed’ is displayed, try reloading by pressing ‘F5’ button), choose ‘Benjamini Hochberg’ for the ‘Test Correction’ and ‘biological_process’ for the ‘DataSet’. Select the top 10 enriched GO terms, click the ‘View results’ button. In the result page, click the ‘Save as List’ then select ‘Create list’ and click ‘Gene’ on the pull-down menu. Name the list as ‘Top 10 GOBP’ and click ‘Create’ to create a list.
- In the Disease Enrichment widget, choose ‘Benjamini Hochberg’ for the ‘Test Correction’ and ‘All’ for the ‘DataSet’. Select the top 10 enriched Disease terms, click the ‘View results’ button. In the result page, click the ‘Save as List’ then select ‘Create list’ and click ‘Gene’ on the pull-down menu. Name the list as ‘Top 10 Disease’ and click ‘Create LIst’ to create the list.
15. Then you will get 3 new lists.