 TargetMine Auxiliary Toolkit is a suite of programmes that features interactive data analysis tools to query and analyse the biological data compiled within the TargetMine data warehouse. TargetMine users can use this toolkit to discover new hypotheses interactively by performing complicated searches with no programming and obtaine the results in an easy to comprehend output format. The auxiliary toolkit enhances TargetMine currently in two ways: 1) biological enrichment analysis with automatic data conversion, graphs and charts and 2) composite interaction network. These functions can be accessed from List pages in TargetMine or via designated input forms (see below)
TargetMine Auxiliary Toolkit is a suite of programmes that features interactive data analysis tools to query and analyse the biological data compiled within the TargetMine data warehouse. TargetMine users can use this toolkit to discover new hypotheses interactively by performing complicated searches with no programming and obtaine the results in an easy to comprehend output format. The auxiliary toolkit enhances TargetMine currently in two ways: 1) biological enrichment analysis with automatic data conversion, graphs and charts and 2) composite interaction network. These functions can be accessed from List pages in TargetMine or via designated input forms (see below)
 Biological Enrichment Analysis
Biological Enrichment Analysis
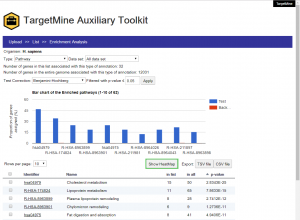
The users can upload a list of genes, extend the query list by including PPI partners and perform orthologue conversion for subsequent enrichment analysis, with only a few clicks of the mouse. Clicking the “Show HeatMap” tab will enable the users to view the selected gene-biological theme annotations as a heatmap. The enrichment analysis results are returned as a table and a histogram that display the distribution of the enriched biological themes in the query list vis-à-vis the background (whole genome annotations). The user may sort the output table (and the histogram) by p-values or the number of annotated genes (foreground or background) or the biological theme names. In the case of KEGG pathway enrichment analysis, the user may click on the pathway name in the output table and the associated genes are automatically sent to the KEGG website and highlighted on the corresponding pathway map.
[Biological Enrichment Analysis]
 Composite Interaction Network
Composite Interaction Network
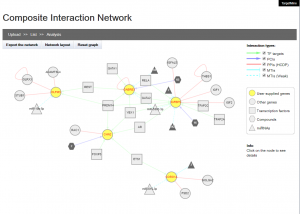
Users can supply a list of genes to construct and visualise a composite interaction network that includes all the biomolecular interactions within TargetMine, i.e, PPIs, MTIs, PCIs/drug-target interactions and TF-target gene interactions, which are associated with the query genes. In a composite interaction network, the different biomolecular interaction types are illustrated in different symbols and colours and the users can choose a variety of layouts (such as force-directed, circular and organic) and also display/undisplay specific interaction types. The users may also filter PCIs (ChEMBL only) by selecting a desired activity threshold
[Composite Interaction Network]
Enrichment analysis web service
A web service for biological theme enrichment analysis. No account or login is required.



