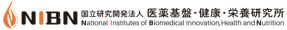Introduction
Gene-set-functional-enrichment (GSFE) relies on a statistical analysis of the relative abundance of biological themes, e.g. pathways, associated with a given gene set and identifies themes (and associated genes) that are overrepresented and therefore, likely to be more relevant to the biological conditions under study. However, the available pathway resources often differ widely in scope and content, which severely hampers a unified analysis and interpretation of high-throughput biological data using diverse pathway repositories. Integration of pathway repositories offers significantly attractive benefits in terms of more extensive and robust functional annotations, which in turn will contribute to a better understanding of gene function and regulation in complex biological systems. Furthermore, it also lends itself to providing a more concise and relatively discrete representation of enriched biological themes in combined GSFE studies.
Traditional gene-set-functional-enrichment (GSFE) study …
Too many and possibly redundant biological themes
New approach …
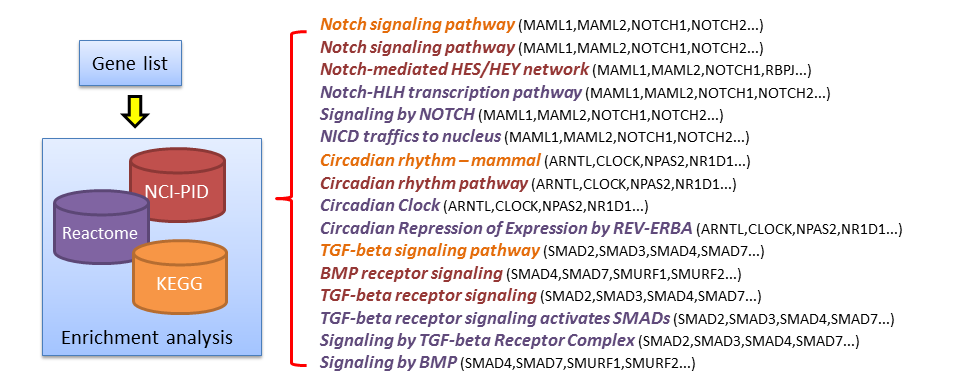
A smaller and distinct list of biological themes with informative labels
Methods
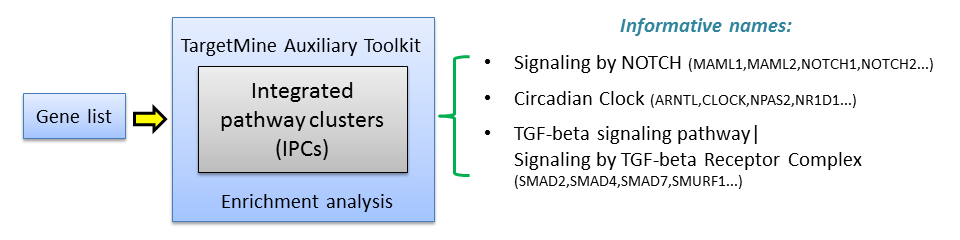
We have proposed a method for pathway clustering base on shared gene content, on the premise that significant overlaps in gene content between the pathways should reflect overall functional congruity between them. An outline of our approach to integrating pathway data from KEGG, Reactome and NCI-PID databases is shown below.
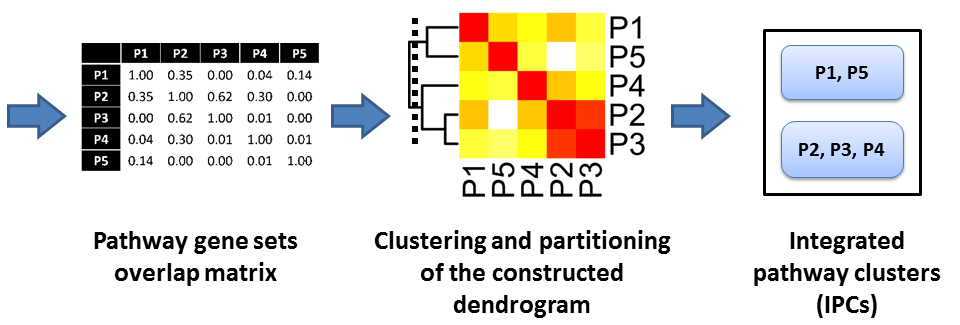
Validation
Functional and Biological Relevance of IPCs
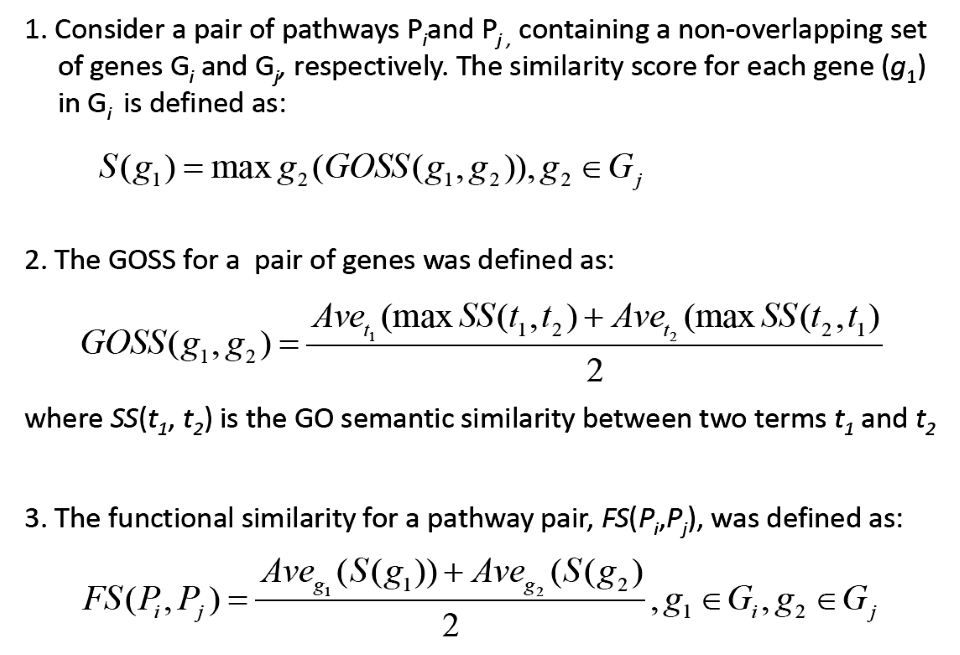
The results have be confirmed by using Gene Ontology Semantic similarity (GOSS) to estimate the functional similarity (FS). GOSS is defined as the extent of relatedness between two GO terms based on the similarity in their annotations; we employed the method of Wang et al. (G-SESAME). The approach is described as the belows.
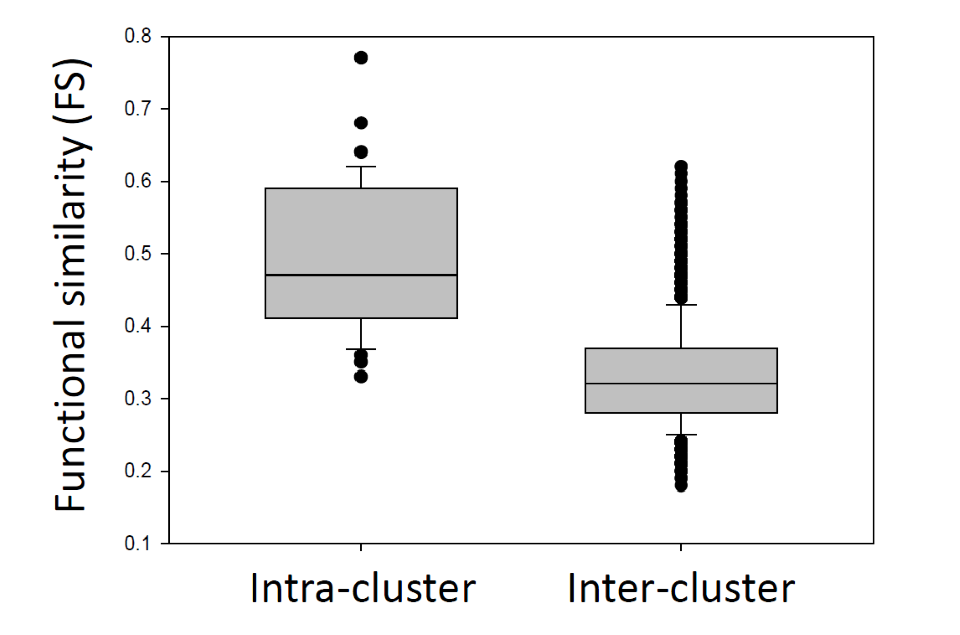
IPCs included pathways, which shared an overall higher functional similarity with each other (intra-cluster) than with pathways from different clusters (inter-cluster). (p = 2.2×10-16)
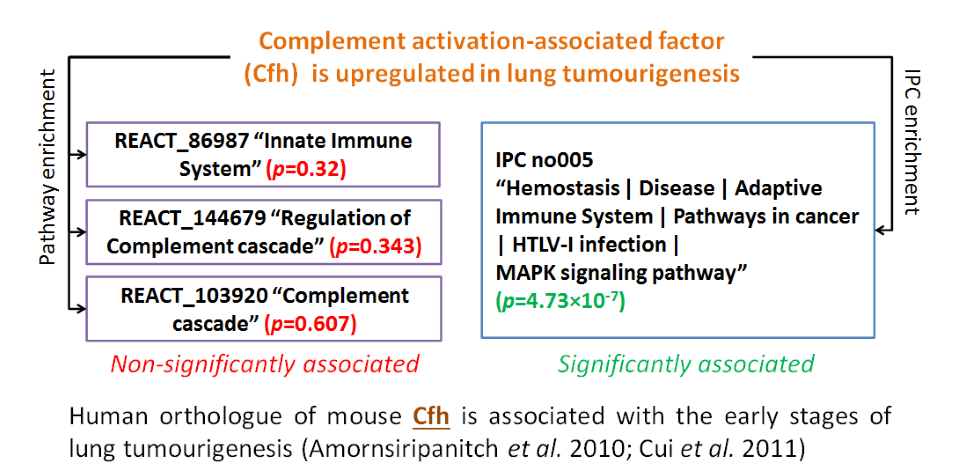
Assessing IPCs in target prioritisation – GSFE analysis
A list of differentially expressed genes from a study of carcinogen-induced lung tumourigenesis in mice (Lokesh et al. 2012), was analysed using both pathways and IPCs. IPCs pick up associations which may often be overlooked by standard analysis.
The IPCs are integrated in TargetMine including the Auxiliary Toolkit. More details and biological applications could be found in the original paper.
Reference:
- Chen Y-A, Tripathi LP, Dessailly BH, Nyström-Persson J, Ahmad S, Mizuguchi K. (2014) Integrated pathway clusters with coherent biological themes for target prioritisation. PLoS One. 9(6):e99030. [PubMed:24918583]