We will describe how to create compound-disease relationship dataset in the following two case.
- 1 Via Disease (Gene/DisGeNET)
- 2 Via Gene Disease Pair (ClinVar, GWAS and dbSNP)
The dataset includes the gene-disease associations inferred from genetic variations.
Case 1.1 Via Disease (Gene/DisGeNET)
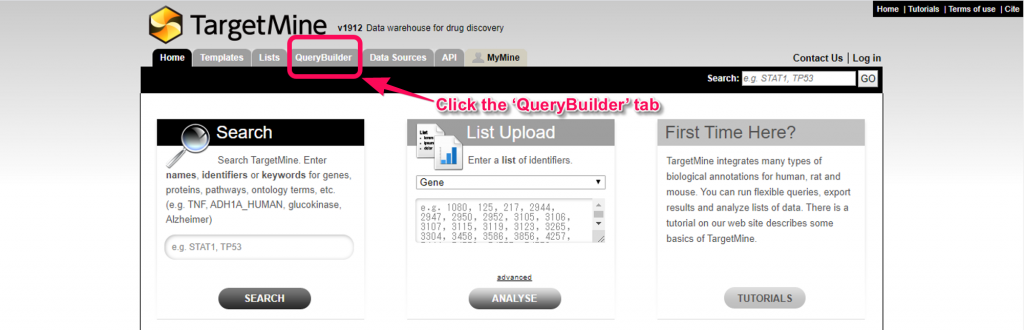
1. Go to the TargetMine main page and click tha ‘Query Builder’ tab.
2. Select the ‘ChEMBL Compound’ and click the ‘Select’ button to begin a query.
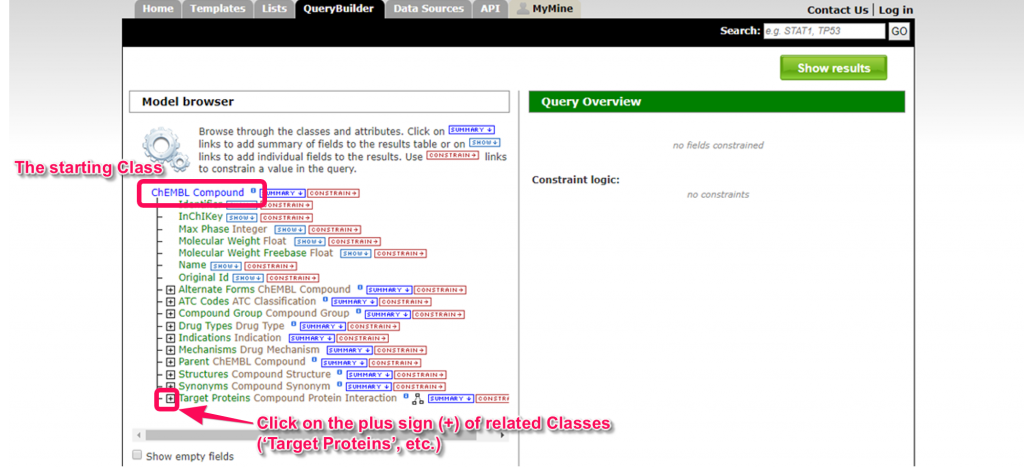
3. The ‘Model browser’ displays TargetMine data model in a tree structure. If the user clicks on the plus sign (+) next to the class name, a list of sub classes is displayed.
For example, if we start from the ‘ChEMBL Compound’ root class, and click next to ‘Target Proteins’, the sub classes ‘Activities’, ‘Data Set’ and ‘Protein’ will be displayed.
To reach ‘Disease Term’, you have to go from ‘ChEMBL Compound’, ‘Target Proteins’, ‘Protein’, ‘Genes’, ‘Gene Disease Pairs’, and finally, ‘Disease Term’.
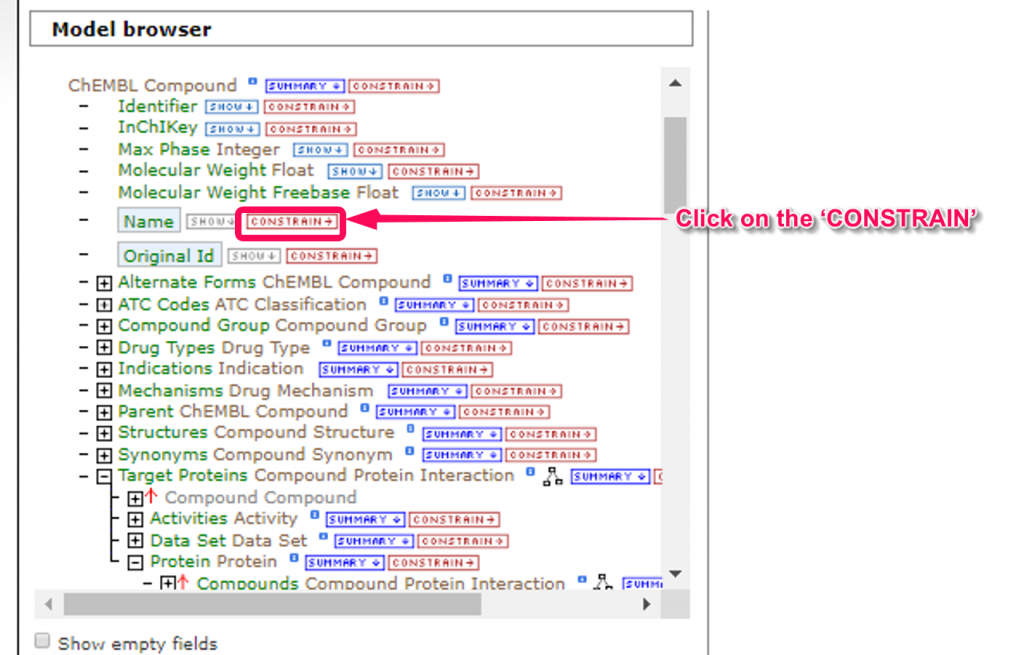
4. You can select the fields to add the result table by clicking on the ‘SHOW’ link. For example, let’s select ‘Name’ and ‘Original Id’ in ‘ChEMBL Compound’ class, ‘DB identifier’ and ‘Symbol’ in ‘Genes’ class, and ‘Identifier’ and ‘Name’ in ‘Disease Term’ class.
Selected fields are displayed in the ‘Query Overview’.
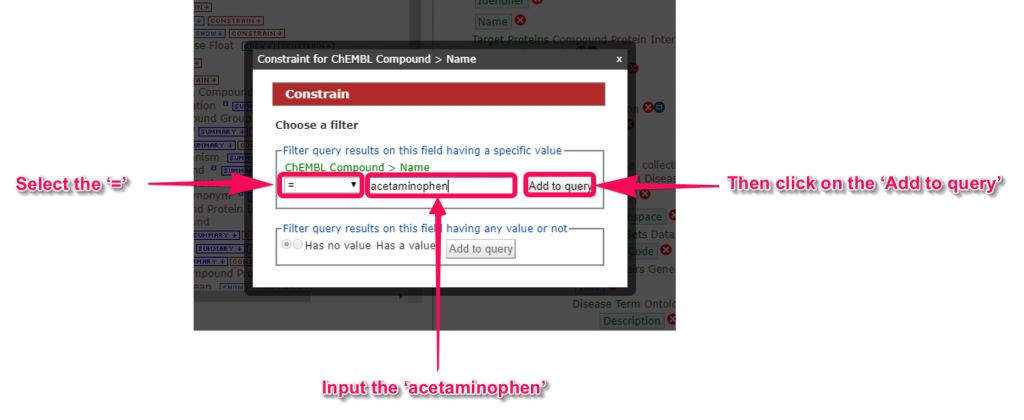
5. In this tutorial, we will put the restrictions on retrieval range in the ’ChEMBL Compound’ for immediate results. When we click on the ‘CONSTRAIN’ link of ‘Name’ in ‘ChEMBL Compound’ class, the ‘Constrain’ window will pop up. For example, you can input the ‘acetaminophen’ and click the ‘Add to query’ button. Unrestricted retrieval will apply a high load to the server, therefore, let us know before submitting your job.
You can download an unrestricted result table from here.
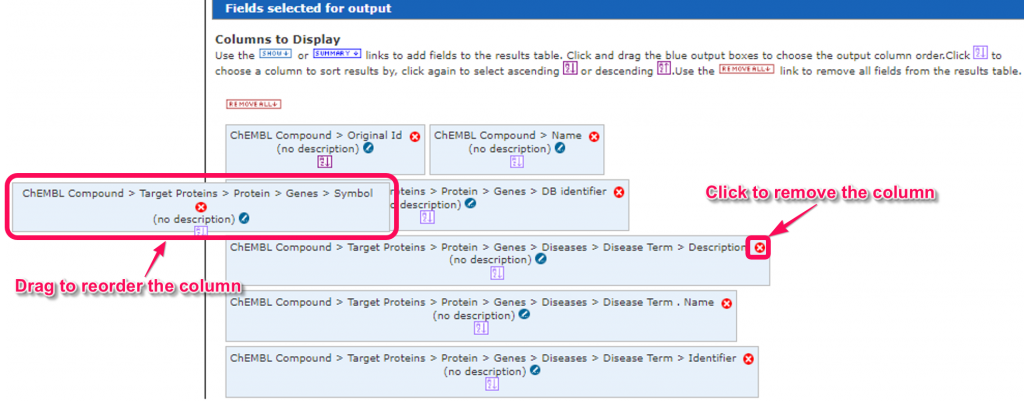
6. In the ‘Fields selected for output’ area, we can change the order of the fields in the column of the results table by drag and drop the field boxes.
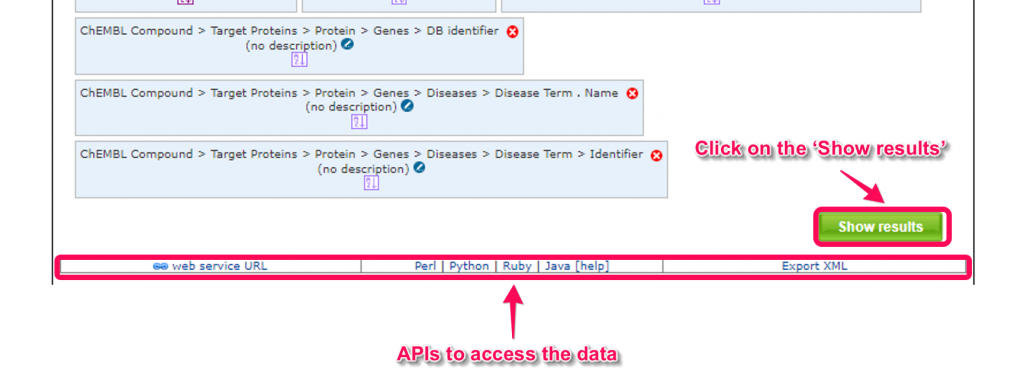
7. Click on the ‘Show results’ button to obtain the query results.
You can also use several kinds of application programming interfaces (APIs) to access the data by HTTP request or programming languages including Perl, Java, Python, Ruby and JavaScript.
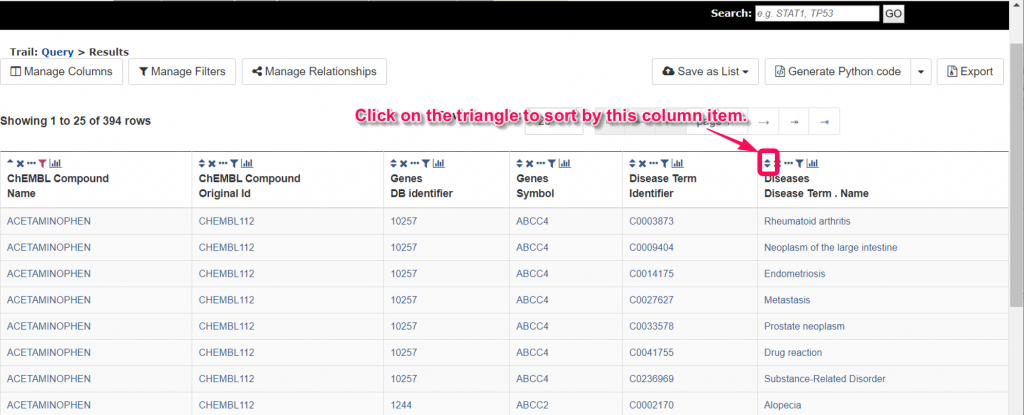
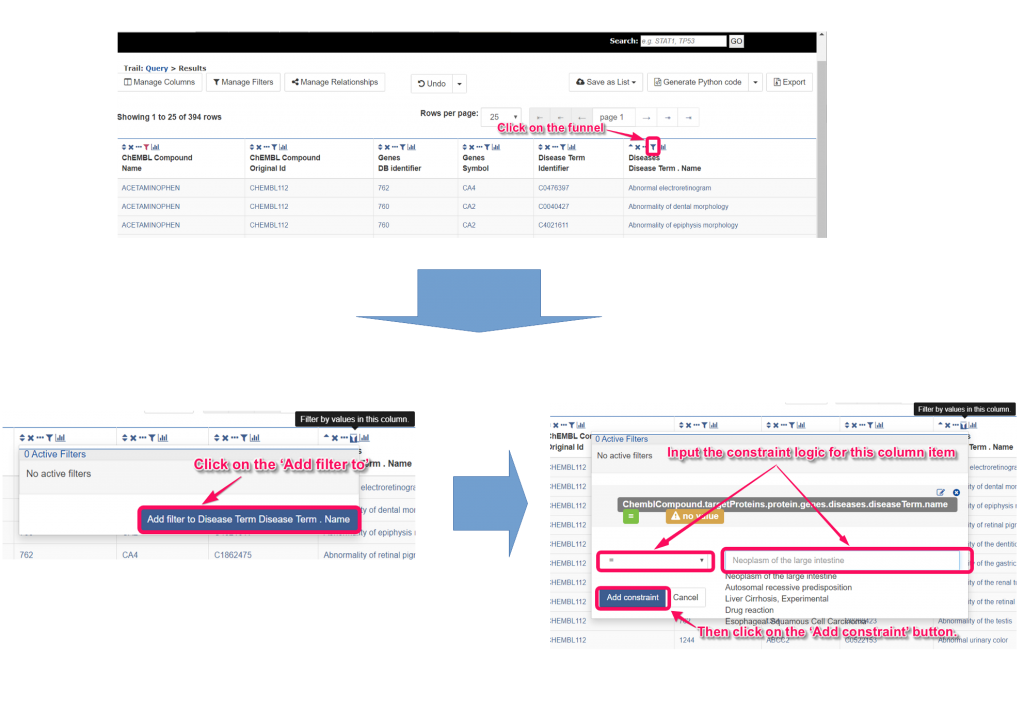
8. We can sort or filter the list by click on the ‘▲’ or ‘▼’ or click on the funnel icons. In the popup panel of the funnel icon, you can click on the ‘Add filter to’ button and input the constraint logic. After clicking on the ‘Add constraint’ button, you can obtain the filtered results.
9. Click on the ‘Export’ button to save the query results.