‘Gene Disease Pair’ compiles gene and disease ontology term associations from different sources.
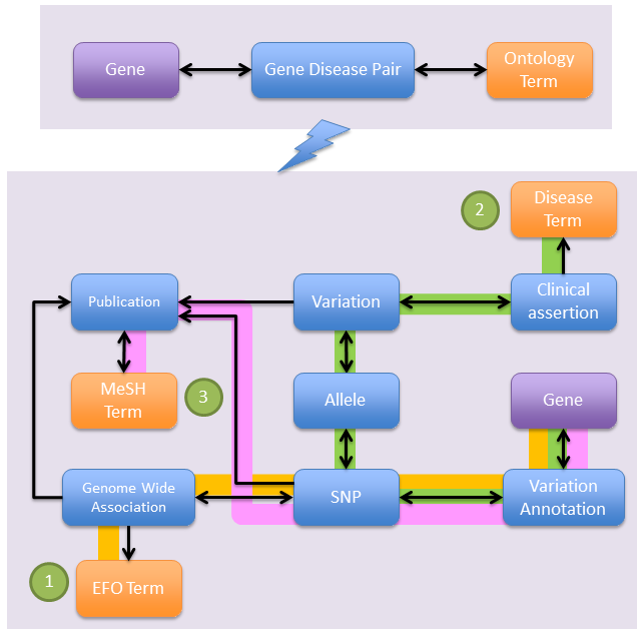
Genes are associated to disease ontology terms; EFO①, MedGen② and MeSH③ through SNP data in GWAS(orange line), ClinVar(Green line) and dbSNP(Pink line), respectively.
Ex. Creating compound-disease dataset based on ClinVar
In Case 1.2, the disease is linked via ClinVar, GWAS, and dbSNP. On the other hand, by adding a restriction to “Disease Term”, it can be related only by one of the sources.
For example, to create a compound-disease dataset based on only ClinVar:
Do after step5 of Case1.2. When we click on the ‘CONSTRAIN’ link of ‘Disease Term’ class, the ‘Constrain’ window will pop up. You can select the ‘Disease Term’ in pulldown menu of ‘Constrained to be in subclass’ after click on the caption ‘Filter query results based on this field being a member of a specific class of objects’. Finally click on the ‘Add to query’ button.
![Correspondence between subclasses of disease terms and limited sources [Subclass] - [source] 'Disease Term' - 'Clin Var' 'EOF Term' - 'GWAS' 'MeSH Term' - 'dbSNP'](https://targetmine.mizuguchilab.org/wp51/wp-content/uploads/2020/03/GeneDiseasePairClass-8-14-724x1024.png)
Correspondence between subclasses of disease terms and limited sources
[Subclass] – [source]
‘Disease Term’ – ‘ClinVar’
‘EOF Term’ – ‘GWAS’
‘MeSH Term’ – ‘dbSNP’
To see and save the query result, please see step 8 and 9 in case 1.1.