TargetMine has been used for data analysis in the following studies:
- HCV pathogenesis (yeast two-hybrid assays): Tripathi et al. 2010.
- HCV pathogenesis (proteomic data): Tripathi et al. 2012.
- Lung tumorigenesis (gene expression): Ihara et al. 2012.
- HCV pathogenesis (yeast two-hybrid assays and literature): Tripathi et al. 2013.
- Cancer immunotherapy (single-cell gene-expression profiles): Nakae, et al. 2015.
- Alzheimer’s disease (genome-wide siRNA screening): Camargo, et al. 2015.
- Colitis (microRNA): Lee et al. 2016.
- Idiopathic interstitial pneumonias (proteomic data): Hamano et al. 2017.
- COPD and IPF (gene expression): Tripathi et al. 2020.
Here are outlines of some of these studies:
Example 1
click on the figure for details.
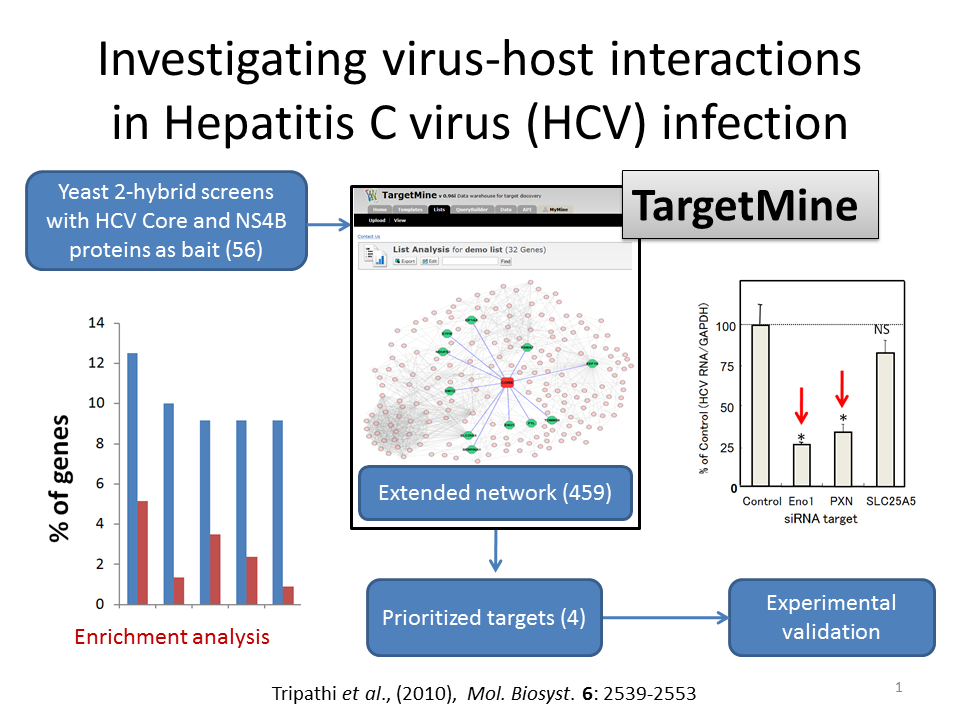
Fifty-six proteins were identified by yeast 2-hybrid screening. The proteins were combined with their interacting partners to construct the extended network. The four prioritized targets are enolase (ENO1), paxillin (PXN), SLC25A5 and vinculin (VCL).
Example 2
click on the figure for details.
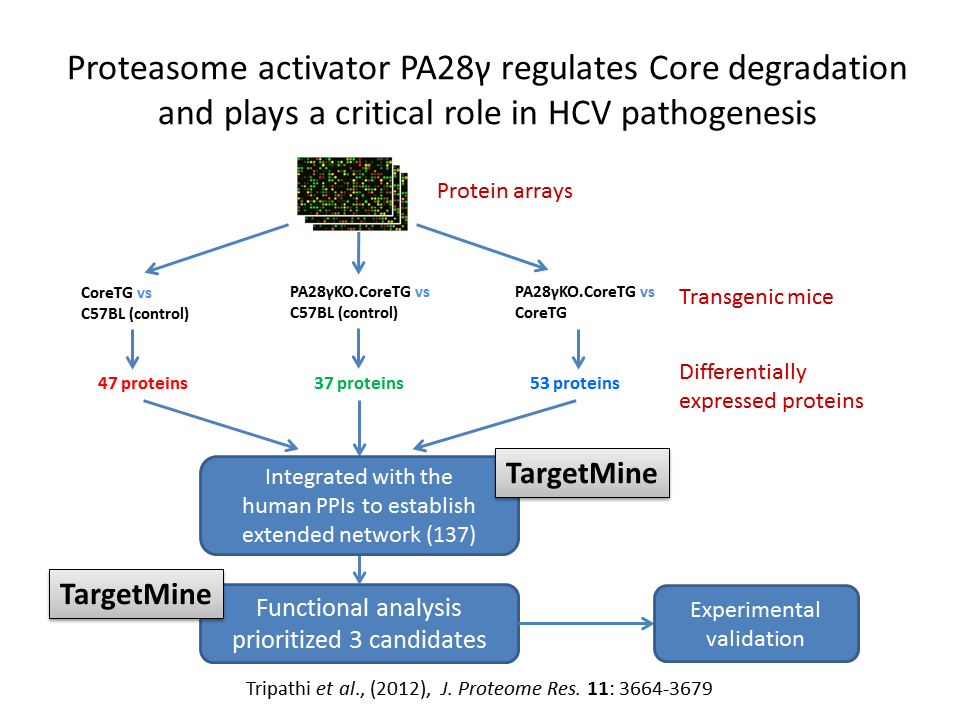
In order to understand the roles of PA28-gamma in HCV infection, we compared and investigated the levels of cellular proteins in transgenic mice expressing HCV core protein in the liver (an in vivo model of HCV pathogenesis) With or without knock out of PA28-gamma expression.
Example 3
click on the figure for details.
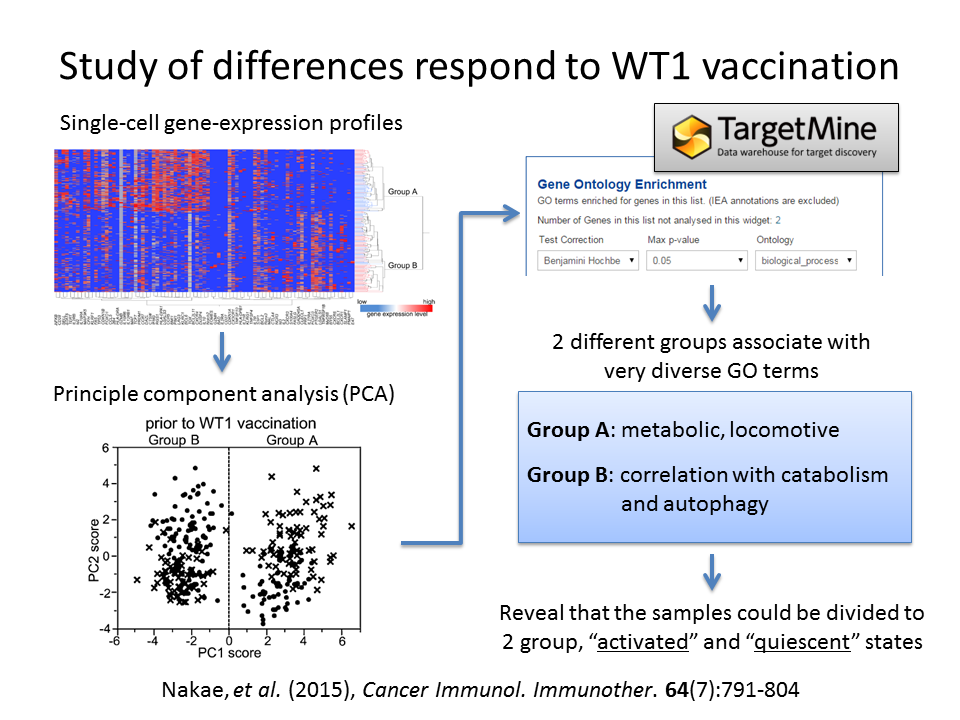
83 genes of the single-cell gene-expression profiles were divided to 2 different groups by the principle component analysis (PCA). Genes that positively contributed to inclusion of single cells in each group were identified by the PC1 loading value and then performed a Gene Ontology (GO) enrichment analysis theme. The 2 groups were found to be enriched for divers GO terms and revealed that the existence of 2 different states, “activated” and “quiescent”.
References:
- Tripathi LP, Itoh MN, Takeda Y, Tsujino K, Kondo Y, Kumanogoh A and Mizuguchi K (2020) Integrative Analysis Reveals Common and Unique Roles of Tetraspanins in Fibrosis and Emphysema. Front. Genet. 11:585998. [doi: 10.3389/fgene.2020.585998]
- Hamano Y, et al. (2017) Classification of idiopathic interstitial pneumonias using anti–myxovirus resistance-protein 1 autoantibody. Sci Rep 7, 43201. [https://doi.org/10.1038/srep43201]
- Lee J, et al. (2016) Profiles of microRNA networks in intestinal epithelial cells in a mouse model of colitis. Sci Rep 5, 18174. [https://doi.org/10.1038/srep18174]
- Nakae Y, et al. (2015) Two distinct effector memory cell populations of WT1 (Wilms’ tumor gene 1)-specific cytotoxic T lymphocytes in acute myeloid leukemia patients. Cancer Immunol Immunother. 64(7):791-804. [PubMed:25835542]
- Camargo LM, et al. (2015) Pathway-based analysis of genome-wide siRNA screens reveals the regulatory landscape of APP processing. Plos ONE 10(2):e0115369. [PubMed:25723573]
- Tripathi LP, et al. (2013) Understanding the Biological Context of NS5A-Host Interactions in HCV Infection: A Network-Based Approach. J Proteome Res. [PubMed:23682656]
- Ihara S, et al. (2012) Inhibitory Roles of Signal Transducer and Activator of Transcription 3 in Antitumor Immunity during Carcinogen-Induced Lung Tumorigenesis. Cancer Res. 72(12):2990-2999. [PubMed:22659452]
- Tripathi LP, et al. (2012) Proteomic Analysis of Hepatitis C Virus (HCV) Core Protein Transfection and Host Regulator PA28γ Knockout in HCV Pathogenesis: A Network-Based Study. J Proteome Res. 11: 3664-3679. [PubMed:22646850]
- Tripathi LP, et al. (2010) Network based analysis of hepatitis C virus Core and NS4B protein interactions. Mol Biosyst 6: 2539-2553. [PubMed:20953506 ]