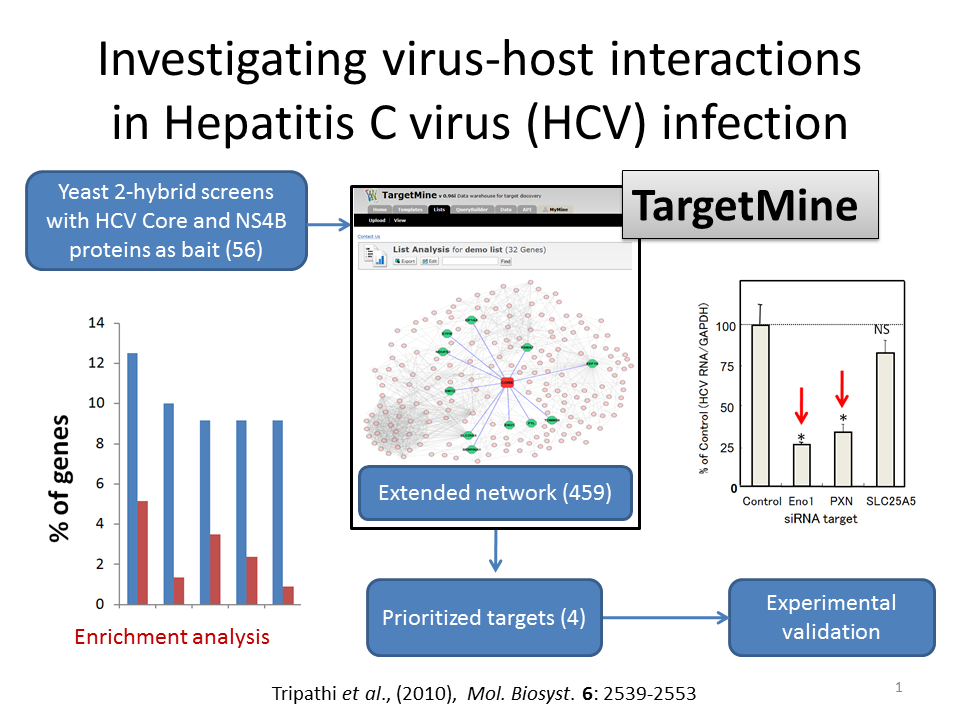
 To understand HCV-host interactions in HCV pathogenesis, a series of yeast-2-hybrid (Y2H) screens were employed, which led to the identification of 56 human host proteins interacting with Core and NS4B (11 interactors for Core and 45 interactors for NS4B). The Y2H interactions were integrated with the human interactome to infer extended PPI networks with the help of TargetMine, to further understand the biological significance of Core and NS4B interactions in HCV infection. The extended PPI networks were then investigated for enrichment of specific biological associations (KEGG pathway, GO biological processes and OMIM phenotypes). Observations of the enriched biological themes and the associated genes in the extended networks suggested that four out of 459 candidates were putative targets for HCV replication and persistence in the host.
To understand HCV-host interactions in HCV pathogenesis, a series of yeast-2-hybrid (Y2H) screens were employed, which led to the identification of 56 human host proteins interacting with Core and NS4B (11 interactors for Core and 45 interactors for NS4B). The Y2H interactions were integrated with the human interactome to infer extended PPI networks with the help of TargetMine, to further understand the biological significance of Core and NS4B interactions in HCV infection. The extended PPI networks were then investigated for enrichment of specific biological associations (KEGG pathway, GO biological processes and OMIM phenotypes). Observations of the enriched biological themes and the associated genes in the extended networks suggested that four out of 459 candidates were putative targets for HCV replication and persistence in the host.
The four targets, enolase (ENO1), paxillin (PXN), SLC25A5 and vinculin (VCL), were assessed for their possible function in HCV replication and release by cellular assay using siRNA. ENO1 siRNA knockdown was found to inhibit HCV replication in HCV genotypes 1b and 2a and viral RNA release in genotype 2a. PXN siRNA inhibition was observed to inhibit replication specifically in genotype 1b but not in genotype 2a, while SLC25A5 siRNA facilitated a minor increase in the viral RNA release in genotype 2a. These results suggested that at least two of four prioritized genes were novel potential drug targets for suppressing HCV infection.
Reference:
- Tripathi LP, et al. (2010) Network based analysis of hepatitis C virus Core and NS4B protein interactions. Mol Biosyst 6: 2539-2553. [PubMed:20953506 ]