[Creation of the compound-disease dataset as examples of AI datasets]
A compound-disease relationship dataset is useful for drug repositioning. In TargetMine, compounds can be connected to diseases by exploring many different relationships including inferred associations.
We will describe how to create compound-disease relationship dataset in the following examples.
- Case 1. Based on known or inferred gene-disease relationships
- Case 2. Based on the co-occurrence in the publication
Comparison of datasets for drug repositioning
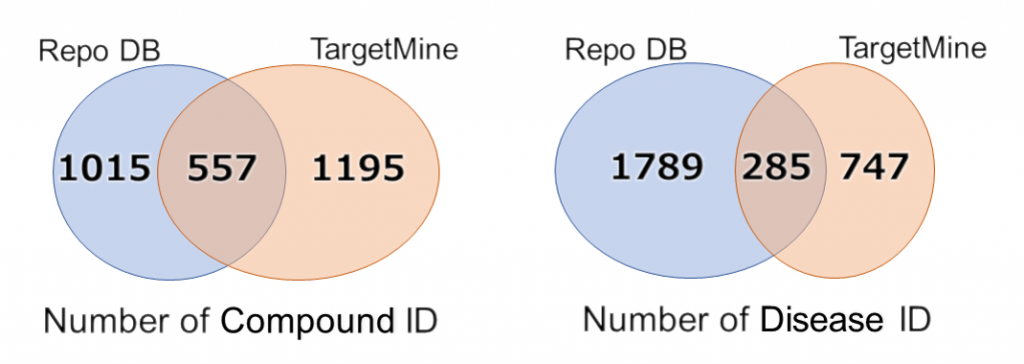
The figure describes a comparison of the compound-disease relationships in the two datasets; the dataset obtained from repoDB[1] and the dataset from TargetMine[2].
[1]Drug Repositioning Database
Sci Data. 2017 Mar 14;4+170029. doi: 10.1038/sdata.2017.29.
Spans:
– drugs: 1,571
– UMLS disease concepts: 2,051
Accounting for:
– approved: 6,677
– failed drug-indication pairs: 4,123
Data points:
– 10,562
[2]Integrated dataware house
(You can create datasets from various DBs by specifying conditions.)
PLoS One. 2011 Mar 8;6(3):e17844. doi: 10.1371/journal.pone.0017844.
Use internal release version as of September 2019.
Spans:
(The datasets using DrugBank Interaction.)
– compound ID: 1,962 (source: DrugBank or KEGG)
– disease ID: 477 (source: MeSH) / 1,031 (source: UMLS)
Data Points:
– 74,454