The ‘List’ function in TargetMine enables queries on user-supplied list(s) of objects (e.g., a list of genes/proteins). The user may either save lists from results pages or create them by uploading lists of identifiers. Lists can be used to constrain queries – either a template query or in the query builder, to perform logical operations (union, intersect, subtract) and exported. The users may also search and copy the lists.
- View a list
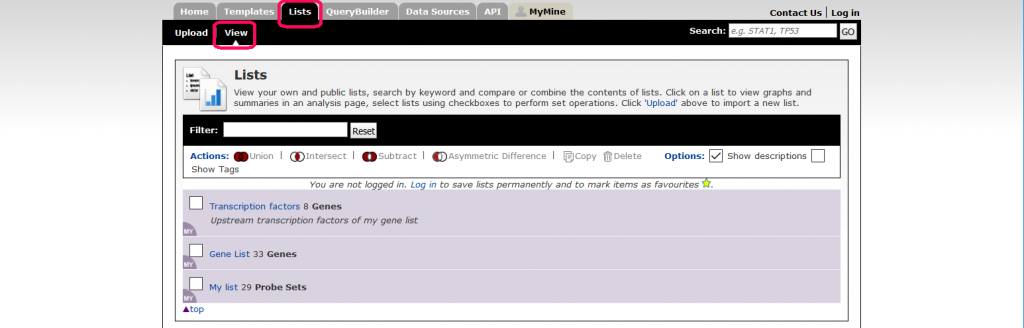
The Lists page is accessible via the ‘Lists’ tab on the TargetMine menu bar. You can view your existing lists in the ‘View’ tab.
- Upload a list
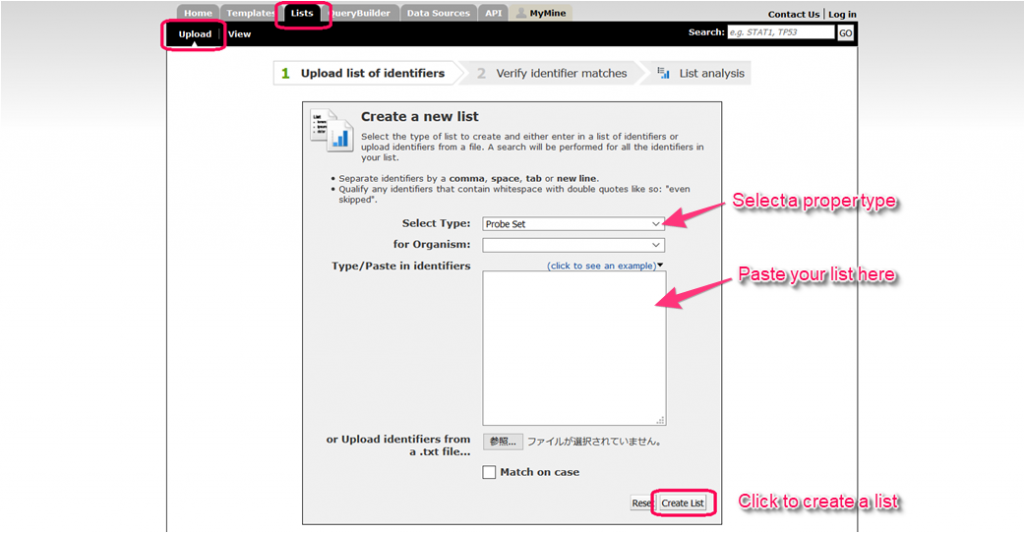
The ‘Upload‘ tab in the Lists page permits the user to create a list of objects either by ‘Type/Paste in identifiers’ or upload a file containing a list of objects.
A list can be created of only one ‘type’ of objects (e.g., genes, proteins), which can be set from the drop-down list. However, it is possible for the list to contain a mix of identifier types (e.g., gene symbols and gene IDs). Optionally, the user may also set the organism for the list of identifiers.
- List Confirmation
Before a list is created it is checked against the TargetMine database and any discrepancies are reported in the ‘No matches found’. The user may either assign a name to the list or use the default name generated by the system.
- List Analysis Page
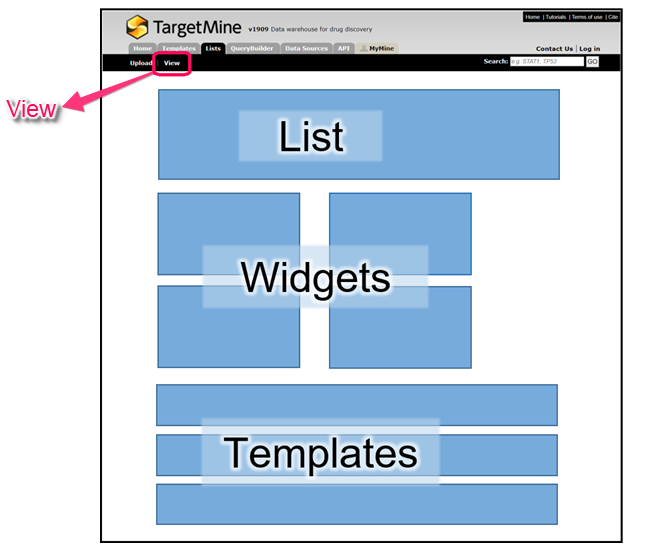
Details of the contents of the lists together with additional information about a list can be viewed through the ‘List Analysis’ page. The analysis page is automatically displayed when a list is created. It can also be accessed at other times by visiting the ‘View’ in the ‘List’ tab, then clicking on the list name.
-
- A summary of the contents of the list includes a table with the objects of the list, a list description, List Info, Convert tools and Link outs (if available).
- A series of ‘widgets’ providing more information about the list, for example, overrepresented KEGG pathways, Gene Ontology (GO) terms, compounds and Disease Ontology associations and CATH classifications.
- A set of template queries that have been pre-run on the set of objects in the list.